Former Research Projects
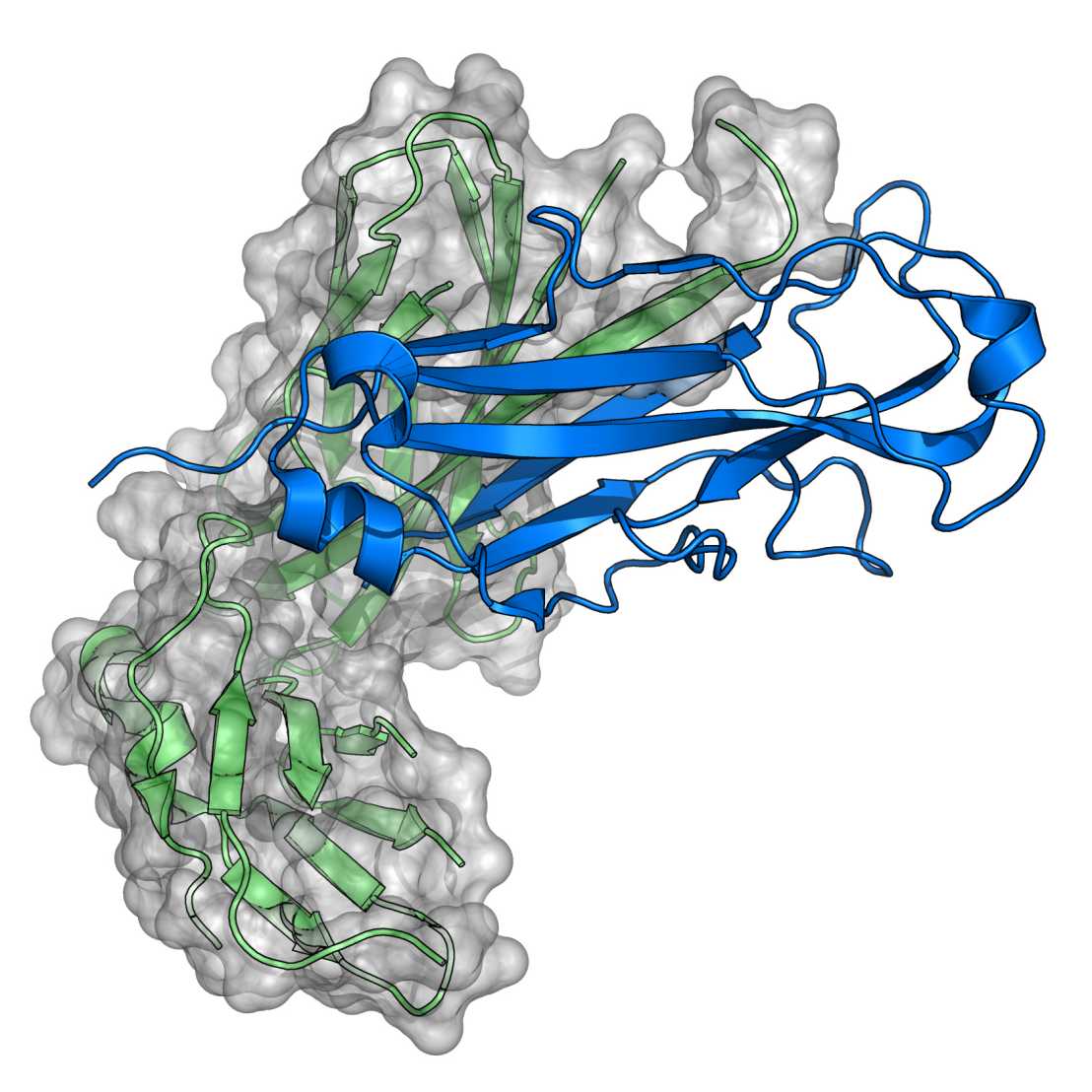
The three-dimensional structure of a protein is exclusively determined by its amino acid sequence and is formed in an autonomous and spontaneous process. Compared to protein folding in vitro, the kinetics and the yields of folding in the living cell are significantly increased by enzymes catalyzing rate-limiting folding steps or preventing nonproductive folding reactions.
An important part of our work is focused on the characterization of bacterial enzymes catalyzing disulfide bond formation during folding of secretory proteins. Our goal is the elucidation of general principles underlying the function of these enzymes and their interactions with folding polypeptides.
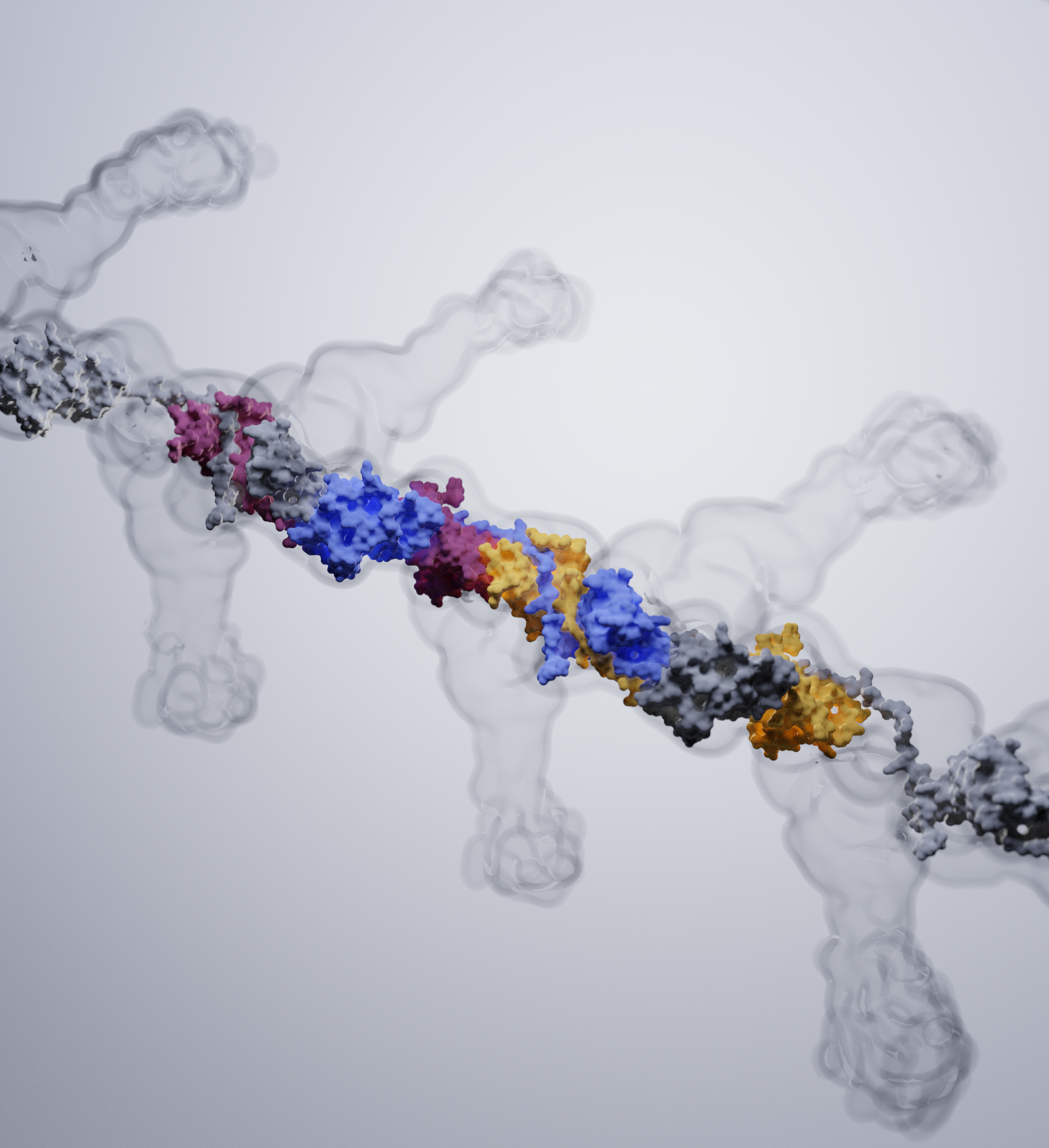

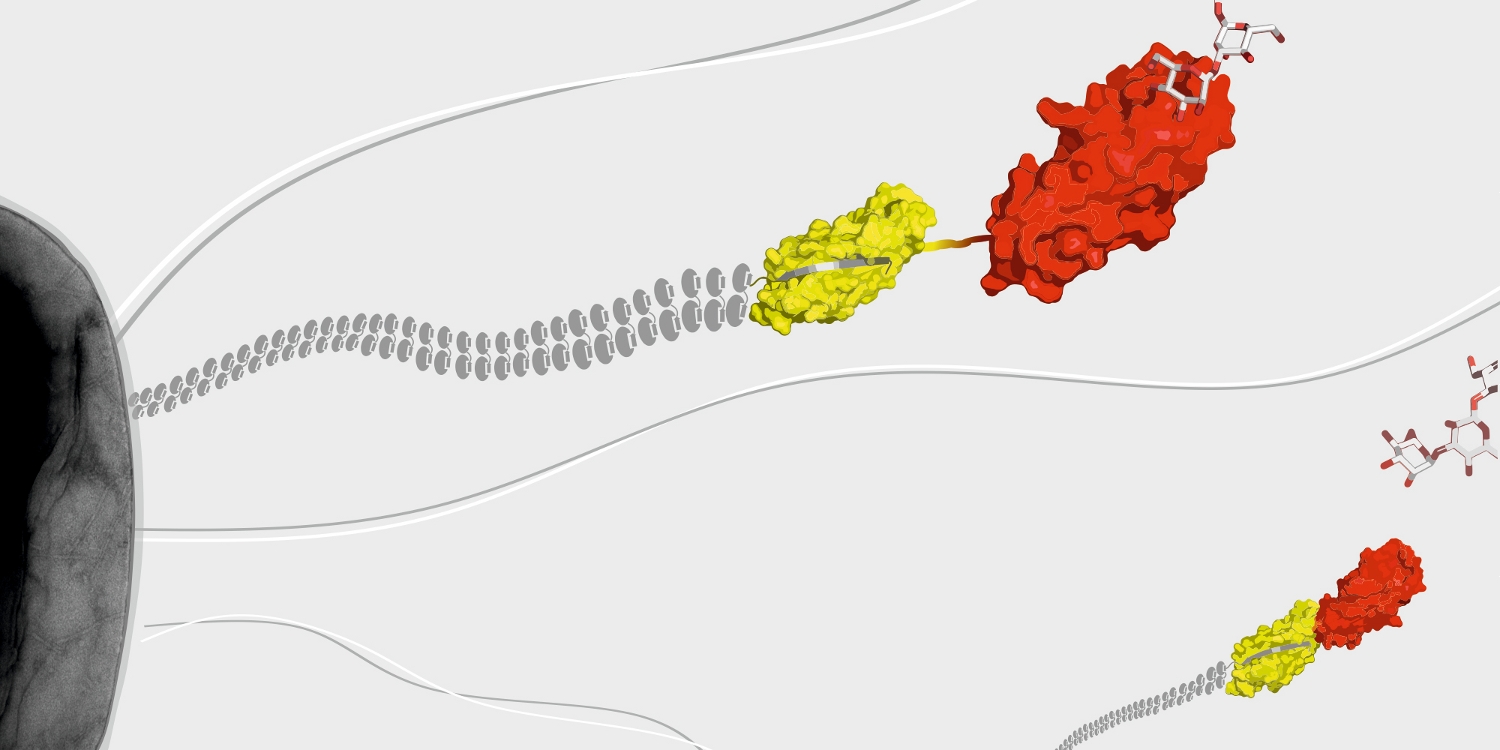
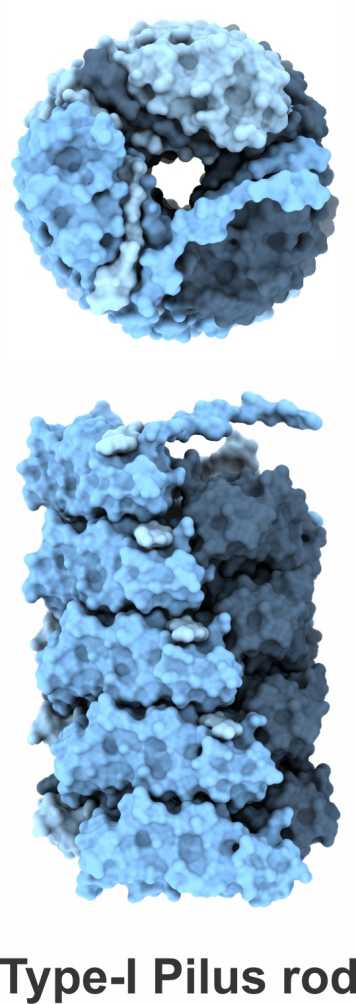
A further research topic is the mechanism of the assembly of adhesive type-1 pili from E. coli. Type-1 pili are large, heterooligomeric protein filaments of uropathogenic E. coli strains that are required for the attachment of the bacteria to host cell surfaces. We are particularly interested in the function of FimC, a periplasmic assembly factor which is not a structural component of the pili but required for pilus assembly in vivo.
Finally, we are trying to answer general questions on protein folding where we focus on random mutagensis techniques in conjunction with screening or selection procedures at the bacterial colony level.
Structure gallery
All our structures can be found on the external page Protein Data Bank website